Day 2 :
Keynote Forum
Alexander A Zamyatnin
A.N. Bach Institute of Biochemistry, Russia; Universidad Técnica Federico Santa Maria, Chile
Keynote: Fragments, fragmentome and fragmentomics in proteomics
Time : 09:30-10:00

Biography:
Alexander A Zamyatnin is a Physicist at M V Lomonosov Moscow State University. He has completed his PhD in Physics and Mathematics (Biophysics), DSci degree in Biology (Human and Animal Physiology) and Professor title in Biophysics. He worked at different scientific organizations of several countries (Russia, USA, Hungaria, Chile, etc.). He has studied “Thermodynamics of muscle contraction; creation of the EROP-Moscow oligopeptide database; computer biophysics and biochemistry of structure and functions of the natural regulatory oligopeptides”. He has more than 200 scientific publications, participated in the national (Russian and Chilean) scientific projects.
Abstract:
Natural fragmentation of biological molecules is well known. Fragmentary structural organization is characteristic of both the simplest and most complex biological molecules. Low molecular weight fragments of biological substances can be easily seen on metabolic maps. Therefore, the term “fragmentomics” is grounded and defined, the bases and determination are given for the notion of the “fragmentome” as a set of all fragments of a single substance as well as for global fragmentome of all chemical components of living organisms. A steady increase in the number of publications dealing with protein fragment has been seen in recent years. For some proteins, there are already hundreds of fragments that have been studied in detail, and it seems that concepts concerning functional importance of the totality of possible fragments of a single protein will be formed. For peptide structures, fragmentomics can be considered as a notion that combines proteomics and peptidomics. EROP-Moscow (Endogenous Regulatory Oligo Peptides) database demonstrates structural and functional variety of possible protein fragments. Formation of an exogenous-endogenous pool of oligopeptides in an organism and correlation of these data with concepts of structure-functional continuum of regulatory molecules is shown on an example of milk and meet protein fragments.
- Track 4: Bioinformatics Algorithms & Databases
Track 5: Immunology and Drug Discovery
Track 6: Perspectives of Clinical Informatics

Chair
Nabil Mili
University of Geneva, Switzerland

Co-Chair
Tamas Hegedus
Semmelweis University, Hungary
Session Introduction
Maria Vittoria Cubellis
University of Naples, Italy
Title: Workshop on Disease specific databases for personalized medicine: Fabry disease as a case of study
Time : 14:20-15:05

Biography:
Maria Vittoria Cubellis graduated in Chemistry from the University of Naples and obtained the PhD in Biochemistry. She is professor of Biochemistry and Bioinformatics at the University "FedericoII" of Naples. Her recent research interests are focused on pharmacological chaperones for the treatment of rare diseases and in particular on the prediction of mutations associated to Fabry disease which are responsive to drugs. She is also trying to extend the approach with pharmacological chaperones to other pathologies, such as disorder of glycosylation type 1a, a disease for which there is no cure at present.
Abstract:
Next generation sequencing of all exons will become common also for symptomless people in the near future. Since we all have approximately 20000 variants, differentiation among non-pathological, mild or deleterious mutations will be necessary. We will show how disease specific databases and predictive tools can be precious for personalized diagnosis and therapy. The analysis of sequence conservation among orthologous proteins, even in the absence of structural information on the human protein, can be sufficient to identify severely pathological mutations, but training sets represent the weak point in the development of prediction tools. At present more than 70000 missense mutations have been reported, with 7 variants per protein on average, but at least 70 cases more than 100 variants are known. A quantitative phenotype can be associated to mutants measuring the residual activity or the stability of proteins expressed by transient transfection. In this case it can be possible to predict the severity of the mutation quantitatively. This implies that in many cases it is possible to develop disease specific predictive tools. Lysosomal alpha galactosidase, which is associated to Fabry disease, represents a good case to test the effectiveness of specific tools that allow predictions scored according to severity. More than 380 missense mutations are known and for 305 the residual activity in cells has been assessed. For the deficiency of lysosomal alpha galactosidase, disease specific predictive tools can also be exploited to estimate responsiveness to specific drugs such as pharmacological chaperones.
Nabil Mili
University of Geneva, Switzerland
Title: Translating genomic biomarker findings into clinical medecine: Dimension of the problem, network structure of competing models and estimate of the misclassification prediction error
Time : 15:05-15:25

Biography:
Nabil Mili completed his Doctorate in Medicine and MSc in Statistics from the University of Geneva. He is board certified in Anesthesiology (Switzerland). His domains of interest are “Model selection in high dimensional data and the connection between statistical formalization and medical reasoning”. He is currently working in the Research Center for Statistics-University of Geneva.
Abstract:
Technical breakthroughs have enabled unprecedented progress in the field of omics. Arguably, this should result in great potential in the field of biomarker discovery and indeed, publications in the field of biomarker discovery have increased dramatically over the past two decades. However, the increase in the number of clinically useful biomarkers have been meager. The major statistical challenges in the translation from biomarker discovery to clinical utility, as long as the concern is to classify any patient into a nosographic category are: The dimension of the problem (how many biomarkers do we need to properly classify a patient?); the network structure of the selected biomarkers (a biomarker does not act in an isolated way but is inserted into a network. What is the architecture of that network?); the paradigmatic structure of competing statistical models (many rival models may have the same misclassification predictive error. They should be considered as equivalent or belonging to the same paradigmatic class) and; an estimate of the misclassification prediction error. We applied a newly proposed gene selection method based on statistical and machine-learning principles which delivered a set of models that best predicted the disease class. These models were inserted in a network where the biomarkers were placed in specific positions according to their relevance in discriminating between the diseases. The principles of the method that meets the above mentioned challenges will be presented and applied to clinical cases (inflammatory bowel diseases, breast cancer and gliomas).
Patrick Kück
The Natural History Museum London, UK
Title: PhyQuart-A new algorithm to avoid systematic bias & phylogenetic incongruence: Are directed quartets the key for more reliable supertrees?
Time : 15:25-15:45

Biography:
Patrick Kück has completed his PhD at the Zoological Research Museum A. Koenig and the Rheinische Friedrich-Wilhelm University in Bonn, Germany, with research focus on the development of new algorithms for data evaluation and homology assessment in phylogenetic reconstructions. He is currently an IEF Research Fellow at the Natural History Museum London, UK. He has published more than 20 papers in reputed journals.
Abstract:
Recent phylogenetic studies of old taxonomic relationships point out how sensitive probabilistic tree reconstruction methods are to the selection of model assumptions and data compositions. Systematic errors are characterized by getting increasingly apparent, the more data are analyzed. An alternative to phylogenetic reconstruction of complete data sets is the divide and conquer principle which divides overall reconstruction problems into smaller subsets. The phylogenetic information gained from such subset analyses is subsequently used to generate a phylogenetic supertree comprising all taxa. Quartet based methods are very attractive for supertree reconstructions. A quartet topology comprises the phylogenetic information inferred from a set of four taxa sequences and is the smallest phylogenetic informative unrooted tree. This talk presents PhyQuart, a new algorithm based on a site pattern classification for quartets of aligned sequences using observed and expected split-supporting site-patterns, considering two different topological directive transformations for the inner branch of each quartet relationship. Simulation analyses show that the combination of site pattern- and Maximum Likelihood analysis leads to quartet inferences that are nearly as good or in many cases even better than in conventional ML-analyses, especially with strong rate heterogeneity. First tests in combination with a new developed supertree technique suggest that PhyQuart might be a good alternative to reduce systematic bias in quartet-based divide and conquer approaches.
Alvaro Olivera-Nappa
University of Chile, Chile
Title: VHL-Hunter: A predictor of the impact of missense mutations in the function of the Von Hippel-Lindau protein (pVHL), risk of clear cell renal carcinoma and severity of pVHL-associated diseases
Time : 15:45-16:05

Biography:
Alvaro Olivera-Nappa has received a PhD in Chemical Engineering, specializing in Biotechnology and Protein Engineering. He was a Post-doctoral Fellow in Santiago, Chile, and Delft, The Netherlands, and has occupied a Visiting Scholar position at the Department of Biochemistry of the University of Cambridge, UK, under Professor Sir Tom Blundell. Since 2013, he is an Assistant Professor at the University of Chile. He has published articles in the fields of protein science, protein engineering, mathematical modeling of biological systems and the prediction of the effect of mutations in cancer and disease. He has developed mathematical, statistical and computational tools to understand and design protein function from a molecular and relational point of view, to represent and analyze complex chemical and biological processes and to design and implement biotechnological solutions and applications. He is interested in biotechnology applications for economic development and constantly promotes innovation and science education as drivers of social development.
Abstract:
Von Hippel-Lindau (VHL) disease is an autosomal dominant syndrome associated with multiple tumors including hemangioblastoma, clear cell renal carcinoma (ccRCC) and pheochromocytoma (PCC), which results from mutations in the VHL gene. VHL disease is classified into type 1 or type 2 depending on the presence or absence of PCC. A major limitation is that accurate classification can only be made in large kindreds. Furthermore, its use in assisting clinical management is limited since a family may move from one subtype to another. More than 600 VHL disease mutations are missense, which are broadly distributed throughout the gene. Inheritance of VHL mutations in an autosomal recessive fashion can lead to congenital polycythaemias. Germline VHL mutations account for up to 50% of patients with apparently isolated familial PCC and 11% of patients with an apparently sporadic PCC. Additionally, somatic biallelic inactivation of VHL also occurs in the majority of sporadic ccRCCs, with ~250 different missense mutations described. Numerous studies have investigated with conflicting results whether partial/total loss of VHL function or the type of VHL mutation may influence prognosis in ccRCCs. Our group previously developed symphony, a predictor of ccRCC risk associated to VHL mutations. In this work we present VHL-Hunter, a new computational approach to predict all VHL-associated disease risks, which combines structural and H-bond network induced pVHL amino acid partitioning, MOSST and SDM predictions. VHL-Hunter correctly predicts the disease outcome of known pVHL mutations with high accuracy (0.96) and informedness (0.9), and also the outcome of previously undescribed mutations. VHL-Hunter also gives clues regarding the mechanism underlying disease generation and is able to correlate the predicted degree of protein malfunction with the severity of clinical phenotypes, in agreement with experimental data. This method can also be successfully applied to obtain highly accurate predictors of mutation phenotypic outcomes for other proteins
Runsheng Chen
Institute of Biophysics, Chinese Academy of Sciences, China
Title: Big Data in noncoding RNA and precision medicine
Time : 16:05-16:25

Biography:
Runsheng Chen, Principal Investigator at Institute of Biophysics, CAS, is an Academician of the Chinese Academy of Sciences (CAS) and an Academician of the International Eurasian Academy of Sciences. He is a member of Human Genome Organization (HUGO), a member of the biomacromolecule group of the Committee on Data for Science and Technology (CODATA) and a member of the bioinformatics professional committee of the International Union of Pure and Applied Physics (IUPAP). He is now the General Secretary and Vice President of Chinese Society of Biophysics and has published more than 130 papers in SCI.
Abstract:
The living organisms on the earth from prokaryotes to eukaryotes have been proliferating for billions of years. To date, they form in more complicated structure and function in more perfect ways. However, what really determined the complex phenotype, structure and function of living organisms? Where do they store those huge amounts of information? And how do they operate? All of these have been keen questions for people to explore. What has been astonishing and puzzle is the fact that life is not just a simple group of molecules instead, it is highly organized. There is connections between nucleus and cytoplasm, a clearly work division between different organizations and synergy cooperation within organs. Therefore, a normal living organism is extremely multi-level and dynamic. The complexity of the organism is not only reflected in the complexity of the structure of DNA information but also on the implementation of the information and operation rule. This report mainly introduces the rise of noncoding area, great innovation opportunity it offers and the role of big data in this field. Meanwhile, this report also introduces what the scientists has been explored for the associations between genotype and phenotype. As a result, series of new concepts such as translational medicine, personalized medicine and precision medicine etc. have been put forward by medical scientists. All of these imply that the big changes for medical system from diagnosis and treatment to health care are upcoming. It also suggests the birth of a new generation of huge health care industry.
Tamas Hegedus
Semmelweis University, Hungary
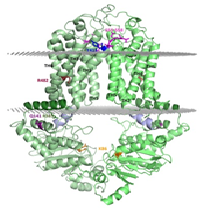
Title: In silico modeling of ABCG2/BCRP structure and dynamics sheds light on xenobiotic recognition and transport function
Time : 16:45-17:05

Biography:
Tamas Hegedus has been a graduate student in the laboratory of Balazs Sarkadi (NIHI, Budapest, Hungary) studying ABC proteins. He has completed his Postdoctoral studies on CFTR/ABCC7 from the laboratory of Jack Riordan (Mayo Clinic Scottsdale, AZ and UNC at Chapel Hill, NC, USA). He leads a group aiming to understand multidrug recognition and folding problem of CFTR mutants (www.hegelab.org). He is a board member of the Hungarian Society for Bioinformatics and a member of the Disease Database Advisory Council, Human Variome Project.
Abstract:
ABCG2 is an ATP binding cassette transporter protein, containing a nucleotide binding domain and a transmembrane region. It functions as a homo-dimer multidrug resistance (MDR) transporter in the plasma membrane, extruding a wide range of molecules from the cell. ABCG2 affects the pharmacokinetics of various drugs and protects the stem cells and cancer (stem) cells from xenobiotic and chemotherapeutic agents. This transporter is present in key pharmacological barriers and drug metabolizing organs. However, ABCG2 is also involved in the transport of endobiotics, e.g. porphyrins and uric acid. Its Q141K variant exhibits decreased functional expression leading to increased drug accumulation and decreased urate secretion. To overcome MDR and correct ABCG2 related pathological phenomena, learning its structural and dynamic properties is a major objective in the field. Still, there has been no reliable molecular model available for this protein, as the published structures of other ABC transporters could not be properly fitted to the ABCG2 topology and experimental data. The recently published high resolution structure of a close homologue, the ABCG5/ABCG8 heterodimer, revealed a new ABC transporter fold, unique for ABCG proteins. We generated a structural model of the ABCG2 homo-dimer based on this fold and confirmed its validity using experimental data. We also performed molecular dynamics simulations and in silico docking to understand the effect of mutations including R482G and Q141K and the mechanism of substrate recognition. The ABCG2 model, we present may have significant impact on understanding drug distribution and toxicity as well as drug development against cancer chemotherapy resistance or gout.
Surya Raj Niraula
B P Koirala Institute of Health Sciences, Nepal
Title: Probability sampling in matched case-control study in drug abuse
Time : 17:05-17:25

Biography:
Surya Raj Niraula has completed his PhD from Tribhuvan University and Post-doctoral from University of Washington, USA. He is the Professor of Biostatistics at B P Koirala Institute of Health Sciences, Nepal. He has published more than five dozen of papers in reputed journals and has been serving as a Statistical Reviewer in many national and international journals. He was awarded with ‘Young Scientist Award’ in 2009, Nepal Bidhyabhusan KA in 2010 and Honored by the President - Constitutional Assembly, 2011. He has presented many conference papers in USA, UK, Korea, Singapore, Thailand, India and Nepal.
Abstract:
Although random sampling is generally considered to be the gold standard for population-based research, the majority of drug abuse research is based on non-random sampling despite the well-known limitations of this kind of sampling. We compared the statistical properties of two surveys of drug abuse in the same community: one using snowball sampling of drug users who then identified “friend controls” and the other using a random sample of non-drug users (controls) who then identified “friend cases”. Models to predict drug abuse based on risk factors were developed for each data set using conditional logistic regression. We compared the precision of each model using bootstrapping method and the predictive properties of each model using receiver operating characteristics (ROC) curves. Analysis of 100 random bootstrap samples drawn from the snowball-sample data set showed a wide variation in the standard errors of the beta coefficients of the predictive model, none of which achieved statistical significance. On the other hand, bootstrap analysis of the random-sample data set showed less variation and did not change the significance of the predictors at the 5% level when compared to the non-bootstrap analysis. Comparison of the area under the ROC curves using the model derived from the random-sample data set was similar when fitted to either data set (0.93 for random-sample data vs. 0.91 for snowball-sample data, p=0.35); however, when the model derived from the snowball-sample data set was fitted to each of the data sets, the areas under the curve were significantly different (0.98 vs. 0.83, p<.001). The proposed method of random sampling of controls appears to be superior from a statistical perspective to snowball sampling and may represent a viable alternative to snowball sampling.
Veronna Marie
University of KwaZulu-Natal, South Africa
Title: Structural implications of protease mutations in antiretroviral treatment exposed patients infected with HIV-1 subtype C
Time : 17:25-17:45

Biography:
Veronna Marie is pursuing her PhD at the HIV Pathogenesis Programme, Nelson R Mandela Medical School in Durban, South Africa. Her current research focuses on “Structural bioinformatics in HIV-1 drug resistance”. The aim of her current work is to “Find mutational pathways in HIV-1 subtype C that can translate into improved medical outcomes for patients failing therapy in resource limited settings”. Her previous research fields included Environmental Biotechnology and Water Research.
Abstract:
The development of drug resistance mutations (DRMs) within protease often results in therapy failure. Currently, limited studies have focused on the structural implications of DRMs in subtype C. The study aim was to assess structural changes within protease that may provide insight into mutational pathways in subtype C. All protease subtype C sequences were retrieved from public databases. After quality assurance, 1912 sequences remained. Resistance associated mutations were identified via the HIVdb algorithm. Phenotypic variation (PV) was calculated using the ConSurf server. Homology models were predicted using Modeller. High PV indicative of drug pressure corresponded to the sheets, 30s loop, core and active site regions of protease. Interestingly, 296 unique combinations of DRMs occurred; the most common was M46I+I54V+V82A (n=27). Combinations of three or more occurred frequently suggesting that these combinations were not random but might have structural implications. M46I+154V+V82A showed greater distances between the mutated residues in comparison to the wild-type (WT). M46I+I54V+V82A+L76V was also common (n=14). V32I, a darunavir mutation directly interacted with L76V and V82A, moving inhibitors away from I84 and towards I50. I54L, also a darunavir mutation, increased the distance of the hydrogen bonds between the mutant and I47 to twice (4.286 Å) that of the WT (I54+I47), modifying flap flexibility and active site dynamics. The interaction of L76V with darunavir mutations V32I, I54L and I84V may have implications when switching to darunavir after lopinavir failure. Understanding DRM pathways in protease can help to prevent therapy failure and promote drug design for next generation protease inhibitors.
Prakruthi Appaiah
CSIR-Central Food Technological and Research Institute, India
Title: Designer protein enriched with large neutral amino acids: A new approach for treating phenylketonuria
Time : 17:45-18:05

Biography:
Prakruthi Appaiah has completed her MSc in Microbiology (2010) from University of Mysore and currently pursuing her PhD (Life Science) in CSIR-Central Food Technological Research Institute, Mysore, India. After her Post-graduation, she worked as Project Assistant (2011) in Lipid Science and Traditional Food Department, CSIR-CFTRI, Mysore, during which she has published three papers in reputed journals and won Best Oral Presentation Award in the National Conference on Functional Foods in Health & Well-being, Bangalore, India. She has presented a poster in 22nd Indian Convention of Food Scientists and Technologists (ICFOST-2012), organized by AFSTI at CSIR-CFTRI, Mysore, India.
Abstract:
Phenylketonuria (PKU) is a genetically inherited disease caused by the defective phenylalanine hydroxylase (PAH) enzyme. In case of phenylketonuria, the body fails to convert phenylalanine (Phe) to tyrosine (Tyr), resulting in the elevated blood Phe level and consequent neurological damage. Of all therapies, large neutral amino acid (LNAA) supplementation has emerged as a promising approach for the dietary treatment of PKU. The LNAAs compete with Phe for the same L-type LNAA transporter (LAT1, SLC7A5) across the blood-brain barrier, decreasing brain Phe level. Thus, the aim of this study was to design an easily digestible protein enriched with LNAA (except Phe) in accordance with WHO/FAO/UNU specification by homology modeling using αs1 casein as template. The challenge was to maximize the LNAAs content (except Phe) in the protein model by finding a suitable scaffold (like α-helix) for homology modeling. Out of 63 different protein models designed, protein model-54 was selected for its compact 3D structure with only α-helices, high sequence similarity with template (60.4%) and good in silico digestibility. Different software like, SWISS-MODEL, EXPASY tool, PROFUNC, I-TASSER, RaptorX and SAVeS Server were used for the structure prediction and validation of the designed protein. The structures obtained from tertiary predicting software were visualized by discovery, UCSF chimera and Pymol tools. Based on these evaluations, the protein model-54 was found to be the best and reliable model. The presentation will review the strategies used for homology modeling, secondary structure and tertiary structure prediction and validation of the designed protein and discuss its nutritional significance for PKU treatment.
Omar Hussein Salman
Al Iraqia University, Iraq
Title: Remote patient triage and prioritization in mobile telemedicine services
Time : 18:05-18:25

Biography:
Omar Hussein Salman completed his graduation from Gifted Secondary in Baghdad with Diploma in 2001. Then, he joined Al Nahrain University, Baghdad and completed his BSc in Computer Engineering in 2004. He completed his MSc in Computer Engineering in 2007. In 2008, he joined the Iraqi University in Baghdad. He completed his PhD in Computer Systems Engineering from University Putra Malaysia in 2016 and his research was focused on “Multi-sources data fusion processing in telemedicine”. During his PhD. he got patent and published a research article. His research interest includes “Data processing, algorithms, remote applications, bioinformatics, telemedicine and healthcare services”.
Abstract:
The more the worldwide population gets older, the bigger is the need for technologies to monitor and assist patients in healthcare applications. Consequently, in order to accommodate the increasing number of users, the remote patient monitor one of the issues that telemedicine, mobile technology and Wireless Body Area Networks (WBAN) have to tackle on, and it constitutes the main focus of this research. To provide healthcare services for a huge number of users, the healthcare providers triage the patients. Triaging involves an initial sorting of patients in order to prioritize the most emergency patients and to ensure by providing them the appropriate and rapid healthcare services. This study proposes a framework to improve the remote triaging and remote prioritization processes for the patients who are in places that are far from the ED and with no triage nurse. The proposed framework named Multi Sources Healthcare Architecture (MSHA) considers multi-heterogeneous sources: Sensors (ECG, SpO2 and Blood Pressure) and text-based inputs from mobile and pervasive devices of WBAN. Simulation results based on datasets for different symptoms of heart diseases demonstrate the superiority of MSHA algorithms as compared to benchmark algorithms in terms of triaging and prioritizing patients remotely in healthcare applications.
Afsheen Mushtaque Shah
University of Sindh, Pakistan
Title: Hematological Abnormalities in patients with Active Pulmonary Tuberculosis

Biography:
She has completed her Ph.D at the age of 34 years from Sindh University, Jamshoro, Sindh, Pakistan. Working as teacher since 2002 and presently working as Associate Professor at Institute of Biochemistry, University of Sindh, Jamshoro, Sindh, Pakistan. She has published 14 research papers in research journals.
Abstract:
Mycobacterium Pulmonary Tuberculosis (PTB) is an enduring infectious disease of the world. Pakistan is a developed nation where the prevalence of pulmonary TB increases and graded as 5th mid of 22 TB elevated countries and is the second leading cause of death in the world. As a risk factor of PTB, tobacco smoking has increased substantially over the past three decades, especially in developing countries. Multidrug-resistant TB (MDR-TB) has become a significant public health problem. Abnormalities in hemoglobin, hematocrit levels and Erythrocyte Sedimentation Rate (ESR) are usually disturbed in infectious disease. The aim of this study was to determine the hematological changes in active pulmonary TB patients and compare with normal healthy control subjects. Total of 338 patients of both genders age ranged 20 to 60 years, with positive sign and symptoms of tuberculosis were selected from Institute of Chest Disease, kotri, Sindh, Pakistan, Liaquat University of Medical & Health Sciences, Jamshoro, Sindh, Pakistan, Liaquat University Hospital, Hyderabad, Sindh, Pakistan, & Rajputana Hospital, Hyderabad, Sindh, Pakistan. Blood was collected from each subject and sent to the research laboratory. Informed consent was taken prior to collection of sample. Our study was based on two groups. One was the Refamipicin sensitive PTB patients (RSPTB) and the other was Refampicin Resistant PTB patients (RRPTB) The mean ages of both gender of RSPTB and RRPTB patients were 36.7±14.1 and 34.0±13.9 years. The male and female patients of RSPTB were 94 and 70 while in RRPTB the male and females were 100 and 74. In RRPTB, the hematological parameters including RBC, PCV, MCH, MCHC, WBC N, L, M,E, PLT & ESR (4.2 ± 0.5), (32.6±5.1),(26.8±3.8), (31.0±1.4), (1.3±3.5),(82.4±9.2), (16.2±6.2), (11.6±5.5), (7.1±1.0), (3.4±1.4) & (63.3±31.1) were significantly higher and MCV (73.6±12.4) were significantly lower. While in RSPTB (3.7±0.7), (30.6±4.7), (23.1±4.0), (29.8±3.2), (1.2±4.2), (81.6±6.1), (16.98±7.5), (8.99±6.3), (5.1±1.8), (3.4±1.3), & (42.5±43.2) were significantly lower and MCV were (76.4±11.1), were significantly higher. As compare to control group all the biochemical parameters were statistically significant. It is conclude that hematological abnormities have a significant role in cure of patients suffering from active pulmonary tuberculosis.
Yanhong Gao
Chinese PLA General Hospital, China
Title: Sensitivity of Palb2-null tumor cells to an oxidative stress inducing agent

Biography:
Yanhong Gao has completed her PhD from Chinese Academy of Medical Sciences in 2005. She work as an associate chief physician and associate professor at Department of Clinical Biochemistry of Chinese PLA General Hospital since 2005. She is interested in finding tumor new biomarkers for diagnoses and prognostic from blood by advanced technology methods. She has published more than 25 papers in journals.
Abstract:
PALB2 gene mutations, as the newly discovered breast cancer associated gene, has brought new direction for the prevention and treatment of breast cancer. To better understand the function for PALB2 and whether it can be use for drug, we generated p53-single-null (as control) and Palb2; p53-double-null cell lines from the mouse mammary tumors obtained and we found the Palb2-null tumor cells were hypersensitive to DNA damaging agents in previous study. To explore new ways to selectively kill Palb2-null tumor cells, we tested the potential of targeting oxidative stress in the cells. For this purpose, we chose phenethyl isothiocyanate (PEITC) and L-sulforaphane. We tested the sensitivity of 5 different Palb2-null tumor cell lines and 3 different control lines to the drugs. Cells were seeded in 96 well plates and treated with different concentrations of the two drugs for 72 hr. Then, cell viability was measured by CellTiterGlo® assay. Comparing with L-sulforaphane, we found Palb2-null tumor cells were hypersensitive to PEITC. PEITC,a natural compound that is rich in vegetables such as watercress and broccoli, etc. PEITC has long been known to possess anti-cancer activity, has been extensively studied. According the result, it is raising an tempting prospect of preventing or treating PALB2-associated cancers with the inexpensive drug.
Biography:
João Lídio da Silva Gonçalves Vianez Junior is graduated in Biological Sciences at the UFRJ, MSc in Plant Biotechnology at the UFRJ and PhD in Biological Sciences (Biophysics) at the same institution. Currently is general coordinator of the Center for Technological Innovation of Evandro Chagas Institute (IEC) and coordinator of the Bioinformatics Core. It has experience in assembly, annotation of genomes and transcriptomics data from NGS platforms. He has knowledge in molecular phylogeny of microorganisms. It also has experience in modeling, molecular dynamics and anchoring.
Abstract:
Brazil is facing an unprecedented growth in the number of microcephaly cases in babies. This phenomenon coincided with the recent Zika virus (ZIKV) outbreak in this country. Although the Brazilian Ministry of Health was quick to recognize that ZIKV was probably the cause of microcephaly in newborns, the underlying mechanisms leading to the development of this pathology have not been established. To tackle this problem at the molecular level, we employed whole transcriptome sequencing of human neurospheres derived from neural stem cells exposed to ZIKV isolated in Brazil (Asian genotype). Differential gene expression analysis of control (MOCK) and ZIKV infected neurospheres generated a list of 26 down-regulated and 64 up-regulated genes. Among the up-regulated detected genes, the Cyclin-dependent kinase inhibitor 1A (CDKN1A) and the Glial fibrillary acidic protein gene (GFAP) were found. CDKN1A prevents the activation of the Cyclin E/CDK2 complex, acting as a regulator of cell cycle progression during G1 and GFAP is a known marker of astrocytes. We also observed a decrease in the expression of the neurogenic differentiation 1 gene (NEUROD1), which is directly involved in the neurogenic program.Those findings suggest that ZIKV infection induces cell cycle arrest and inhibits the neuronal differentiation, resulting not only in the reduction of the size, but in a deeper disruption of the normal development of the human brain.

Biography:
Dr. P. Ananda Gopu has received his Ph.D. degree in Biotechnology (subject bioinformatics) from Periyar Maniammai University, India in 2012. He has worked as Research Associate at Bioinformatics Research Centre (BIRC) of Nanyang Technological University, Singapore in 2009.08- 2011.03. Dr. Ananda Gopu has joined as Scientist at Centre for Research and Development PRIST University in 2014 and his goal is several applications in health care involved in understanding complex relationship between sequence, structure, functions and interactions of protein-protein & protein-DNA using computational analysis of biological data. He has 7 papers in peer-reviewed international and national journals to his credit
Abstract:
In the present study, the protein sequences have been preconceived in series of atoms instead of amino acids (AA). Specifically, the stretches of protein sequences have been considered preferably in terms of carbon atoms instead of using an AA-based pattern. Accordingly, we have observed that patterns consisting of the same number of carbons can result in different AA lengths with different numbers of total atoms. The variation of these patterns is characterized by a change in the distribution of large hydrophobic residues (LHRs) (such as phenylalanine, isoleucine, leucine, methionine and valine) with the introduction of higher numbers of small hydrophobic residues (SHRs), such as glycine, alanine, proline and cysteine. Consequently, proteins that have the same number of carbons can have different numbers of AA within their various patterns and thus an increase in their peptide length. Hence, proteins of the same carbon contents may show different patterns with different peptide lengths that may reflect their specific biological functions. Concluding, an atomic level comparison of protein sequences can provide better results than a similar comparison at the residues level which may have potential implications for the prediction of misfolded proteins.
