Day 1 :
Keynote Forum
Petra Perner
Institute of Computer Vision and Applied Computer Sciences, Germany
Keynote: Quantitative measurement of cellular events with image processing and data mining for drug discovery, therapy and system biology
Time : 10:35-11:05

Biography:
Petra Perner (IAPR Fellow) is the director of the Institute of Computer Vision and Applied Computer Sciences IBaI in Leipzig, Germany. She received her Diploma degree in electrical engineering and her PhD degree in computer science for the work on “Data Reduction Methods for Industrial Robots with Direct Teach-in-Programing”. Her habilitation thesis was about “A Methodology for the Development of Knowledge-Based Image-Interpretation Systems". She has been the principal investigator of various national and international research projects. She received several research awards for her research work and has been awarded with 3 business awards for her work on bringing intelligent image interpretation methods and data mining methods into business.
Abstract:
In the rapidly expanding fields of cellular and molecular biology, fluorescence illumination and observation is becoming one of the techniques of choice to study the localization and dynamics of proteins, organelles and other cellular compartments as well as a tracer of intracellular protein trafficking. With this arises the problem of the automatic mass analysis of image information. Image-interpretation systems which generate automatically the desired target statements from the images are important. The continuation of mass image analyses on the basis of the classical procedures leads to investments of proportions that are not feasible. New procedures are therefore required. We will present, new intelligent and automatic image analysis and interpretation procedure, based on our flexible image analysis and interpretation system cell interpret. We will demonstrate it in the application of HEp-2 cell pattern analysis.
- Track 1: Evolutionary & Structural Bioinformatics
Track 2: Emerging Trends in Proteomics & Genomics
Track 3: Frontiers of Systems Biology in Bioinformatics

Chair
Alexandru G Floares
SAIA-Solutions of Artificial Intelligence Applications, Romania

Co-Chair
Elena Papaleo
Danish Cancer Society Research Center, Denmark
Session Introduction
Wenzhong Xiao
Stanford University School of Medicine, USA
Title: Of men and not mice: Comparative genomic analysis of human diseases and mouse models
Time : 16:10-16:30

Biography:
Wenzhong Xiao directs the Computational Genomics lab at Stanford Genome Technology Center, Stanford Medical School and is responsible for solving computational problems in the development of new genomic and proteomic techniques and their applications. He is also the Director of the Immuno-Metabolic Computational Center at Massachusetts General Hospital. At the interface of computation, genomics and medicine, he focuses on “Computational challenges in development and application of omics for diagnosis, prevention and therapy, especially of human immune and metabolic diseases”.
Abstract:
A cornerstone of modern biomedical research is the use of mouse models to explore basic disease mechanisms, evaluate new therapeutic approaches and make decisions to carry new drug candidates forward into clinical trials. However, few of these human trials have shown success. Here, we systematically compared the genomic response from publically available datasets of patients of different acute inflammatory diseases and corresponding murine models; and showed that, although inflammation from different etiologies resulted in highly similar genomic responses in humans, the responses in mouse models correlated poorly with the human disease and also with one another. Among genes changed significantly in humans, the murine orthologs are close to random in matching their human counterparts. Our findings suggest that a disease model shall be carefully (re) examined to see how well it reproduces the human disease at the molecular level because virtually every drug and drug candidate target gene product(s). In addition, our comparisons of trauma patients reveal genomic signature between complicated and uncomplicated outcomes, and specific mediators which serve as predictive biomarkers for the development of targeted treatments at the bedside.
Alexandru G Floares
SAIA-Solutions of Artificial Intelligence Applications, Romania
Title: The impact of functional redundancy on molecular signatures
Time : 16:50-17:10

Biography:
Alexandru G Floares is a Medical Doctor (Neurologist) specialized in artificial intelligence applications in Oncology. Currently, he is the President of the Solutions of Artificial Intelligence Applications Organization and the OncoPredict Company. He completed his Neurology specialization and PhD in Biophysics at University of Medicine and Pharmacy, Iasi, Romania. His research project focus on “Developing non-invasive omics tests for cancer diagnostic, prognostic and treatment response prediction and automation of the mathematical modeling of complex biological networks with ordinary and partial differential equations using artificial intelligence”. He has publications and inventions in the fields above.
Abstract:
Precision medicine’s goals cannot be reached without discovering molecular signatures from omics big data, using machine learning. The prevailing conception is that for a given biomedical condition and class of biomolecules, there should be unique and highly accurate signature. Following the best practice for whet lab determinations and predictive models development, high accuracy (>95%) could often be achieved. However, the uniqueness request contradicts biological systems organization. They are amazingly robust, despite being highly complex. Evolution seems to favor functional redundancy which confers robustness. Until recently, we were searching for single relevant biomarkers. Then, we moved to single list of biomarkers. Now, we should move to multiple equivalent lists and even go beyond lists. Multiple different lists could be the results of different feature selection or modeling algorithms. If they are all highly accurate and relevant for the biomedical condition, they probably reflect functional redundancy. As most algorithms were developed with the goal of finding the smallest highly relevant subset of variables, they precluded the proper exploration of redundancy. That is why; we will have multiple equivalent small lists. Integrating many such lists could give a better representation of the biological reality. We can also go beyond the too simple concept of a single list with molecules increased or decreased in a particular condition. Using decision trees, one can obtain rules-sets containing biomarkers, thresholds and their conditions for each phenotypic class. This vision will be illustrated with our results from analyzing the biggest miRNA NGS data collection in cancer from TCGA. This work was supported by the research grants UEFISCDI PN-II-PT-PCCA-2013-4-1959 INTELCOR and UEFISCDI PN-II-PT-PCCA-2011-3.1-1221 IntelUro, financed by Romanian Ministry of Education and Scientific Research.
Magda Babina
Charité University Medicine Berlin, Germany
Title: The mast cell has much in store for us: A call to bioinformaticians to advance insights into a unique, intriguing but underexplored cell subset of the human body
Time : 17:10-17:30

Biography:
Magda Babina completed her PhD in Biochemistry in 1999 from Free University Berlin, Germany. She has studied Mast Cell Biology throughout her career, with emphasis on MCs in humans. She is a Senior Scientist and group Leader at Charite, Department of Dermatology and Allergy, and has published more than 50 papers. She has been a member of the FANTOM consortium (led by RIKEN in Japan) since 2010, one of the largest life science consortia in the world. This collaboration inspired her to obtain a more comprehensive and complete view of human MCs through systems biology approaches in collaboration with bioinformaticians.
Abstract:
Mast cells (MCs) are best known as effector cells of allergy but suspected to perform a range of other functions. Our knowledge of MCs in humans is seriously limited, as was one message from the collaborative endeavor FANTOM5 which used deep-CAGE sequencing on skin-derived MCs to generate a comprehensive view of their transcriptome. MCs were embedded in the body-spanning atlas, the datasets allowed to directly contrast their molecular signature against ≈200 primary cells. Our work demonstrates that: MCs are unique cellular elements; have no near neighbor; are intensely adaptable and; display transcriptional peculiarities. Our work also demonstrates: Uniqueness: MCs combine “private” with pan-hematopoietic genes supplemented by genes of disparate organs (e.g. neuronal/reproductive); Position: MCs have no close relative in the hematopoietic network being well separated from all other lineages, both by principal component analysis and by pairwise correlation analysis; plasticity: MCs show substantial adaptations regarding transcriptome, protein/mediator expression and functional programs in new microenvironments and; peculiarities: Cells with greatest TF diversity across atlas (893/MC versus 617/average) and many non-annotated transcripts exclusively active in MCs. Encouraged by these findings, novel functionalities of MCs have been uncovered (e.g. active BMP receptor and significance of the retinoid network) but burning questions remain such as “What is the nature of the TF network underlying lineage specification?”, “How do non-annotated transcripts contribute to MC identity?”, “How are genes from unrelated tissues activated in MCs”? Detailed bioinformatics analyses will help identify the most probable interconnections to facilitate further examination by wet-bench biologists.
Elena Papaleo
Danish Cancer Society Research Center, Denmark
Title: Structural communication in transcription factors: How interactions with DNA, mutations or post-translational modifications reshape the interfaces for cofactor recruitment
Time : 17:30-17:50

Biography:
Elena Papaleo completed her PhD in 2006 and Post-doctoral from 2007-2009 at the Department of Biotechnology and Bioscience at the University of Milano-Bicocca (Italy) in the group of Prof. Luca De Gioia and Prof. Piercarlo Fantucci. She was then appointed as Adjunct Professor in Computational Biology at the University of Milano-Bicocca from 2010-2012. Afterwards, she was Senior Post-Doctoral Researcher in the group of Prof. Lindorff-Larsen at the Department of Biology of the University of Copenhagen (Denmark) from 2011-2015. She has been Visiting Researcher at many international institutes including the group of Prof. Salvador Ventura at the Institute of Biotechnology and Biomedicine (IBB, Barcelona, Spain) and the group of Prof. Francesco Luigi Gervasio at the Spanish National Cancer Research Center (CNIO, Madrid, Spain). In August 2015, she joined as Group Leader of the Computational Biology (CBL) Laboratory at the Danish Cancer Society Research Center (Copenhagen, Denmark). She has authored more than 50 scientific papers as main or senior author and she is Academic Editor of PLoS One, Frontiers in Molecular Biosciences (Nature publishing group), PeerJ and Journal of Molecular Graphics and Modelling. The main research of CBL focuses on “Molecular modelling and simulations integrated with experimental data and network theory to the study structure-function relationship in key cancer proteins as well as on the analyses of high-throughput sequencing and omics data from profiling of cancer patients”.
Abstract:
Little is known about the molecular mechanisms related to the conformational changes induced at distal sites in many transcription factors which are often related to cancer disease, such as p53, MZF1 and the family of ARID domains. Thus, my group is focusing on the characterization of their structural dynamics to enrich the knowledge on this important group of regulatory proteins. In particular, we are employing a combined approach that integrates atomistic microsecond molecular dynamics simulations, enhanced sampling techniques, methods inspired by graph theory and cross-validation of the simulated ensembles with NMR data. To relate these properties to protein function, we studied both the free and DNA-bound forms of wild type, mutated and phosphorylated variants of p53, ARID proteins and MZF1. The interaction with DNA not only stabilizes the conformations of the DNA-binding loops, but also strengthens pre-existing paths in the free protein for long-range communication to potential interface for cofactor recruitment. Conformational states of these distal regions that are only a minor population of the free ensemble are promoted by DNA interactions, altering the preferences for certain classes of biological partners and thus influencing the signaling pathways mediated by these proteins. Moreover, mutations or post-translational modifications can also contribute to reshape the population of these interfaces even in domains that are not necessarily involved in DNA-binding.
Lee Wei Yang
National Tsing Hua University School of Life Science, Taiwan
Title: GNM 2.0- A database housing protein dynamics data of >11,000 PDB structures revealing dynamics prerequisites for protein functions and interactions
Time : 17:50-18:10

Biography:
Lee Wei Yang is currently an Associate Professor at Institute of Bioinformatics and Structural Biology, National Tsing Hua University, received his PhD degree in Molecular Genetics and Biochemistry from School of Medicine, University of Pittsburgh (2005) and his Post-doctoral training in University of Tokyo (2006-2009), La Jolla Bioengineering Institute and Department of Chemistry, Harvard University (2010-2011) before joining NTHU in 2011. He has published more than 30 papers in reputed journals and has an H-index of 16.
Abstract:
Proteins function through advantageously utilizing a repertoire of possible modes of intrinsic motions. Understanding such motions, of which the importance has been acknowledged in year 2013’s Nobel Prize in Chemistry, is essential to accurately predict two body interactions, channel gating mechanisms and enzyme catalysis. Here, we present the only dynamics database that houses dynamics data (vibrational normal modes) of protein structures in a size commensurate with Protein Data Bank (PDB). The interface that presents such data is state of the art of its kind. Given the wealth of the data, we are able to find dynamics traits for enzyme active sites. Such traits are later used to predict the locations of enzyme active sites for protein structures of a resolution as low as 20 Å. We further data-mined the database and develop the concept of intrinsic dynamics domains (IDDs), including a domain plane (D-plane) and a domain axis (D-axis). It is found that a protein interacts with another at the interface where D-plane cuts through and forming a near-vertical angle between two intersecting D-axes from the two proteins over a set of 68 protein–protein complexes. The findings are then used to define quantitative criteria to filter out docking decoys unlikely to be native whereby the chance to find near-native hits is doubled. Our results also show that in 95% of the DNA-protein complexes, the DNA is cut through by protein’s D-plane. The dynamics database, GNM 2.0, is made available at http://dyn.life.nthu.edu.tw/gnmdb and IDD website is provided at http://dyn.life.nthu.edu.tw/IDD/IDD.php.
Xiaoyu Yu
University of Pretoria, South Africa
Title: Mathematical modelling of evolution of tetranucleotide usage patterns of whole bacterial genomes to improve phylogenetic inferences
Time : 18:10-18:30

Biography:
Xiaoyu Yu has completed his MSc in Bioinformatics and is currently doing a PhD (2nd year) in Bioinformatics at the University of Pretoria. He has currently no publication with one in working progress and has attended and presented (posters) in many conferences in Europe such as ECCB 2014, ECCB 2015 and VAAM2016.
Abstract:
Nowadays, complete genome sequences of multiple bacteria became readily available for analysis. Current work which uses whole genome based alignment (WGS) approach for phylogenetic and phylogenomic research believe to resolve contradiction between gene based trees, but this approach multiplies the problem in terms of gene annotation, orthology prediction and inadequate alignment of sequences. Therefore one of the most prospective ways for genome comparison and phylogenomic inferences is then based on annotation-and-alignment free genome linguistic approaches, i.e. comparison of oligonucleotide usage patterns (OUP) of genome-scale DNA fragments. Until now, this approach still lacks a reliable evolutionary model to explain the mechanisms and dynamics of changes in OUP which hinders the application of this approach to systematically compare to other well-known methods such as marker genes and/or whole genome sequence based alignment. The aim of the current work is divided into three important topics: i) Comparative analysis of multiple complete genome sequences representing different phylogenetic branches at different taxonomic levels to identify the driving forces of OUP evolution; ii) Analysis of topological incongruences between phylogenetic trees based on orthologous gene alignments, whole genome alignments and alignment free OUP patterns; iii) Improving phylogenetic inference by reconciliation of sequence based and pattern based evolutionary models. The major output of this research is an innovative evolutionary model implemented in a form of a computer program for phylogenetic inferences based on combination of alignment based and alignment free approaches.
Matteo Lo Monte
Institute of Protein Biochemistry - CNR, Italy
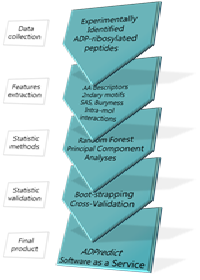
Title: ADPredict: ADP-ribosylation sites prediction based on physicochemical and structure descriptors
Time : 18:30-18:50

Biography:
Matteo Lo Monte completed his PhD in Medicinal Chemistry with a project entitled, “In silico screening of taste receptors: An integrate modeling approach” in 2015, at the “Universita’degliStudi di Milano”, under the supervision of Prof. Giulio Vistoli and in collaboration with Dompé Farmaceutici SpA, under Dr. Andrea Beccari. During this period, he mainly worked on the TRP receptors family, predicting their 3-D structure by homology modeling techniques and studying their interaction capacities as well as their activation mechanisms by Molecular Docking and Molecular Dynamic simulations; so generated models were conveniently utilized in virtual screening campaigns that successfully led to the identification of new hits. He moved to the Institute of Protein Biochemistry at the National Research Council in Naples in 2015 as Postdoctoral Fellow in the research group of Dr. Alberto Luini. He computationally supported several ongoing projects of Molecular Biology through molecular modeling and structural analyses studies of diverse enzymes like β4Galt5, SHIP1 and AGPAT4, as well as investigating the post-translational modifications, ADP-ribosylation in particular, developing predictive algorithms.
Abstract:
Statement of the Problem: ADP-ribosylation is a post-translational modification governing several crucial cellular processes, such as inflammation, cell survival or damaged DNA detection and repairing machinery activation. It is, thus, strictly related to neoplastic conditions. To date, ADP-ribosylation is poorly understood, as still incomplete is the knowledge of its effects on numerous molecular paths. Deeply understanding the circumstances in which it happens, as well as the numerous target proteins and the way their role in their respective biological pathways are affected by this event, would represent an important achievement in molecular biology, not solely for the progression in combating cancer.
Methodology & Theoretical Orientation: ADPredict is an in silico predictive algorithm of ADP-ribosylated Aspartate and Glutamate residues, based on both known physicochemical parameters (Z-Scales, ST-Scales, MSWHIM, ProtFP) and in-house derived secondary structure related and 3-D descriptors of hundreds of human ribosylated proteins. ADPredict was developed using principal component analyses (PCA) and the random forest algorithm. Its predictive capacity was then evaluated via intensive boot-strap approaches.
Findings: Here we present the first computational predictive tool able to individuate the Aspartate or Glutamate residues that are most likely to be ADP-ribosylated in a target of interest. Predictions can be achieved via single or multiple models (meta-model strategy), so allowing each time a tailored approach. It will soon be available as an online service at the website.
Conclusion & Significance: ADPredict arises as a new, concrete support to the study of the ADP-ribosylation event, flanking the analytic approaches developed so far and addressing the experimental investigation of this important biologic phenomenon. Ongoing extension of the predictive algorithm would aim to account also for the modification of additional amino acidic residues, so enlarging the applicability domain of the tool.
Jean-Didier Maréchal
Universitat Autònoma de Barcelona, Spain
Title: GAUDIMM: Widening the application of genetic algorithms for molecular modeling

Biography:
Jean-Didier Maréchal has completed a double PhD in chemistry from the Université Paris Sud (France) and Universitat Autònoma de Barcelona (Spain). He completed four years of Post-doctoral studies in England (Leicester and Manchester) and France (Paris Sud). He returned to Spain in 2006 and now leads his group on molecular modelling and design and presents a record of more than 65 papers in reputed journals.
Abstract:
Here, I present an optimitzation plateform for 3D molecular design based on a multi-objective genetic algorithm. GAUDIMM (Genetic Algorithm Under Design Inference and Molecular Modeling) is able to take several chemical descriptors (genes) at once and optimize them against a number of structural evaluators (objectives). Its aim is to provide with physically sound 3D models and its particularity is to allow the researcher to fill the geometric search with information generally partial provided by different sources, including data from other levels of theory, experiments or chemical intuition. The researcher only has to chose the adequate genes and objectives for his/her problem. GAUDIMM is provided with an API and the users can code their own extensions to adapt the optimization engine to their needs. With the already available built-in descriptors, one is able to select several molecules at once (think about competitive multiligand protein-ligand dockings), explore conformational flexibility (torsion angles and rotamers) and chemical variability (mutated residues, chemical group replacements). On the optimization side, a plethora of objectives have already been implemented from scoring functions to accurate force field energy calculations, simplified chemical interactions (hydrogen bonds, hydrophobic patches, steric clashes, covalent bond formation), geometric evaluation (distances, angles, surfaces areas, volumes, etc..) and metallic coordination. To date, several projects are challenged to tackle by standard computational methodologies have been successful thanks to GAUDIMM and include artificial enzyme re-design, geometries of metal-coordinated peptides and biosensors. While docking is not its main target, our platform even reports good benchmarks when tested against commonly used datasets.
Mahreen Arooj
Curtin University, Australia
Title: Structure-based systems biology for prediction of off-targets, biological pathways & target diseases

Biography:
Dr. Arooj has completed her PhD from Gyeongsang National University, South Korea in 2013. She has worked as Postdoctoral Research Fellow at Plant Molecular Biology and Biotechnology Research Centre (PMBBRC). Currently, she is working as Early Career Research Fellow in the School of Biomedical Sciences, Curtin University, Australia. Her research interests and expertise are to apply computational techniques such as structural bioinformatics, systems biology, and molecular dynamics in the fields of biomedical sciences.
Abstract:
Off-target binding connotes the binding of a small molecule of therapeutic significance to a protein target in addition to the primary target for which it was proposed. Progressively such off-targeting is emerging to be regular practice to reveal side effects. Here,we have developed a robust computational strategy using structure-based systems biology approach that is applicable to any enzyme system and that allows the prediction of drug effects on biological processes. Chymase is an enzyme of hydrolase class that catalyzes hydrolysis of peptide bonds. A link between heart failure and chymase is ascribed, and a chymase inhibitor is in clinical phase II for treatment of heart failure. However, the underlying mechanisms of the off-target effects of human chymase inhibitors are still unclear. In this study, putative off-targets for huamn chymase inhibitors were identified through various structural and functional similarity analyses and molecular docking studies. Finally, literature survey along with KEGG pathway maps was performed to incorporate these off-targets into biological pathways and to establish links between pathways and particular adverse effects. Off-targets of chymase inhibitors are linked to various biological pathways such as classical and lectin pathways of complement system, intrinsic and extrinsic pathways of coagulation cascade, and fibrinolytic system. Prospectively, our approach is helpful not only to better understand the mechanisms of chymase inhibitors but also for drug repurposing exercises to find novel uses for these inhibitors. This study also demonstrates the significance of computational strategies for efficacy prediction and the role that systems biology may play in multitarget therapeutics.
Alexander Zamyatnin
Russian Academy of Sciences, Russian Federation
Title: Fragments, fragmentome and fragmentomics in proteomics

Biography:
Alexander A. Zamyatnin is a physicist originally (M.V.Lomonosov Moscow State University, Physical Faculty), has PhD degree in physics and mathematics (biophysics), DSci degree in biology (human and animal physiology), Professor title (biophysics). Work: in different scientific organizations of several countries (Russia, USA, Hungaria, Chile, etc.). Studies: thermodynamics of muscle contraction; creation of the EROP-Moscow oligopeptide database; computer biophysics and biochemistry of structure and functions of the natural regulatory oligopeptides. He has more than 200 scientific publications, participates in the national (Russian and Chilean) scientific projects.
Abstract:
Natural fragmentation of biological molecules is well known. Fragmentary structural organization is characteristic of both the simplest and most complex biological molecules. Low molecular weight fragments of biological substances can be easily seen on metabolic maps. Therefore the term “fragmentomics” is grounded and defined, the bases and determination are given for the notion of the “fragmentome” as a set of all fragments of a single substance, as well as for global fragmentome of all chemical components of living organisms. A steady increase in the number of publications dealing with protein fragment has been seen in recent years. For some proteins there are already hundreds of fragments that have been studied in detail, and it seems that concepts concerning functional importance of the totality of possible fragments of a single protein will be formed. For peptide structures, fragmentomics can be considered as a notion that combines proteomics and peptidomics. EROP-Moscow (Endogenous Regulatory OligoPeptides) database demonstrates structural and functional variety of possible protein fragments. Formation of an exogenous–endogenous pool of oligopeptides in an organism and correlation of these data with concepts of structure–functional continuum of regulatory molecules is shown on an example of milk and meet protein fragments.

Biography:
Abstract:
Several studies showed the importance of rhizobacteria (P. fluorescens) associated to natural fertilizer improving cereal production in poor and arid soils. However, agriculture crops prediction by statistical models remains an adequate system to assess the performance of bio-fertilizer used in farming practices. Indeed, the large amount of data to process in this context allowed the integration of computers in the statistical analysis schemes. Computational statistic can be defined as the explicit impact of computers on statistical methodology. Here,we developed a bioinformatics pipeline in R bio-statistic environment assessing the relationship between previous analyzed rhizobacteria (P. fluorescens) treatments (T0: treatment without any rhizobacteria and any foliar bio-fertilizer, T1: treatment with only rhizobacteria, T2: treatment with both rhizobacteria and foliar bio-fertilizer and T3: treatment with only foliar bio-fertilizer)and their potential influences on growth and yield parameters of both maize and soybean cereal varieties in arid soil in the north of Côte d'Ivoire. Then, the present survey basing on the computational statistic approach, highlighted a strong difference between the four considered rhizobacteria treatments impacting the two analyzed cereal crops and development process (p-values < 0.05). Moreover, the same analysis suggested a positive and selective effect of rhizobacteria (P. fluorescens) combined with foliar bio-fertilizer on both quantitative and qualitative production of analyzed cereal crop varieties. Indeed, we were able to demonstrate the differences between maize and soybean crops replying torhizobacteria (P. fluorescens) bio-fertilizer treatments. Further, the present developed pipeline showed that the two analyzed varieties of soybean (green and yellow soybean) were differentially influenced by the different rhizobacteria treatments as opposed to maize plant varieties (p-value < 0.05). Finally our findings evidenced that disregarding analyzed parameters and cereal varieties, treatment T2 having the recommended dose of rhizobacteria P. fluorescens+ foliar fertilizer compost recorded the best performance improving both maize (Zea mays. L) and soybean (Glycine max) cereals cultivation in arid region. In conclusion this study demonstrated the key role of rhizobacteria (P. fluorescens) combined with foliar bio-fertilizer improving cereal production in soil with low fertility aptitude, adjusting the concordance between both growth and yield parameters.
