Day 2 :
Keynote Forum
Judit Ovádi
Hungarian Academy of Sciences, Hungary
Keynote: Challenging drug target for moonlighting and chameleon proteins
Time : 09:30-10:00

Biography:
Judit Ovádi has expertise in Biochemistry, Enzymology and Molecular Biology. She defined the metabolic channeling at microscopic and macroscopic levels as a powerful mechanism to control and direct metabolic pathway at crossroads. Her research team showed the sensing potency of the microtubule network at system level that can regulate signaling pathways due to its decoration by interacting partners. They discovered a unique brain-specific protein denoted Tubulin Polymerization Promoting Protein (TPPP/p25) that displays two exciting characteristics: intrinsically unstructured and enriched in brain inclusion in the case of Parkinson disease and other synucleinopathies. The structural and functional features of this protein have been characterized at different levels of organizations under physiological and pathological conditions. She has supervised several DSc and PhD dissertations. She was an invited Visiting Professor in USA, Spain and Italy; Invited Speaker in international congresses in the last few years in Tokyo, San Francisco, Jerusalem, Orleans, Nice. At present, she is a Professor Emerita, Laureata Academiae of the Research Center of Hungarian Academy of Sciences.
Abstract:
The conformational diseases such as Parkinson’s disease (PD) and Multiple System Atrophy (MSA) represent an important group of neurodegenerative disorders. The hallmarks of these diseases are the a-synuclein (SYN) and the recently discovered Tubulin Polymerization Promoting Protein (TPPP/p25). Both proteins are disordered with chameleon characteristics and expressed distinctly in neurons and oligodendrocytes (OLGs), respectively; notwithstanding they are co-enriched and co-localized in pathological inclusions in the case of PD and MSA. TPPP/p25 is the prototype of the Neomorphic Moonlighting Proteins by displaying both physiological and pathological functions due to their interactions with distinct partners. At physiological conditions TPPP/p25 modulates the dynamics and stability of the microtubule system; its expression is crucial for the differentiation of OLGs, the major constituents of the myelin sheath. The assembly of TPPP/p25 and SYN, as fatal initiative, of the etiology of PD and MSA has been established. Due to the unique structural and functional features of TPPP/p25, a new innovative strategy has to be evaluated to inhibit and/or destruct specifically the interaction of TPPP/p25 with SYN; this could be fulfill by targeting of the interface of the pathological complex without affecting the physiological one. Our studies underline that targeting multifunctional proteins is a challenging task; nevertheless, the validation of a drug target can be achieved by identifying the interface of complexes of the partner proteins existing at the given pathological conditions.
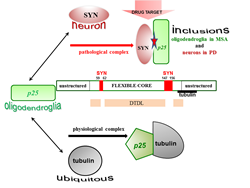
Figure 1: Distinct interfaces of TPPP/p25 are involved in its hetero-association with tubulin and SYN as physiological and pathological partners. The interaction of the disordered TPPP/p25 with tubulin results in significant conformational changes; this effect does not manifest itself in the case of its association with SYN. Although the deletions within the middle CORE segments do not abolish the binding of TPPP/p25 mutants to SYN, except in the case of the double truncated double loop mutant (DTDL), due to its chameleon feature; however, a potential segments (marked red) were identified as potential drug target.
Keynote Forum
Jarrod A Marto
Harvard Medical School, USA
Keynote: CITe-Id as a novel chemoproteomic platform to characterize covalent kinase inhibitors
Time : 10:00-10:30

Biography:
Jarrod A Marto is internationally recognized for his expertise in the development and use of state-of-the-art mass spectrometry and other bioanalytical techniques to characterize cellular communication pathways that underlie normal physiology and human diseases. His lab pursues technology development in mass spectrometry for quantitative analysis of primary human tissues or high-fidelity model systems. He has published widely in the areas of basic chemistry, analytical science, advanced instrumentation, mass/bio-informatics, and cancer biology.
Abstract:
The therapeutic value of targeting protein kinases is demonstrated by the small molecule inhibitors receiving regulatory approval primarily for cancer therapy. Despite these successes, only a handful of truly selective inhibitors have been developed for the nearly 600 human kinases. The recent approval of cysteine-directed covalent inhibitors of BTK and EGFR has reignited interest in covalent kinase therapeutics. One advantage of covalent drugs is their ability to potently and permanently disable protein function often with only transient drug exposure. We are focused on probes which covalently modify members of the cys-kinome, the subset of approximately 200 kinase which harbor a targetable cysteine residue in proximity to the ATP-binding site. We have developed quantitative mass spectrometry approaches which enable site-level interrogation of proteins targeted by irreversible inhibitors on a proteome-wide scale. For individual probes which target kinases such as EGFR, JNK, BMX, FGFR, CDK7, or BTK, we typically identify several hundred intracellular protein targets. We developed a companion, competition-format assay called 'CITe-Id' which discriminates non-specific versus selective, concentration-dependent inhibitor binding to protein targets. Importantly we successfully differentiate the repertoire of binding targets for probes which comprise structurally similar analogs, suggesting an efficient mechanism for medicinal chemistry optimization of second-generation inhibitors. Finally, our quantitative approach provides important clues for development of inhibitors targeting obscure kinases. The combination of structure-guided synthesis informed by CITe-Id chemoformic target and site identification provides a scalable platform that delivers first-in-class covalent chemical probes that may serve as useful starting points for future small molecule therapeutics.
Keynote Forum
Magnus S Magnusson
University of Iceland, Iceland
Keynote: From protein to human cities the first and only large-brain mass-societies: Structural and functional
Time : 10:30-11:00

Biography:
Magnus S Magnusson is a Research Professor. He completed PhD in 1983 from the University of Copenhagen. He is an Author of the T-pattern model initially focused on the real-time organization of behavior has co-directed DNA analysis. He presented numerous papers and invited talks at international mathematical, neuroscience, proteomics, bioinformatics and science of religion conferences and at leading universities in Europe, USA and Japan. He is a Deputy Director during 1983-1988 at Anthropology Laboratory, Museum of Natural History, Paris and repeatedly invited temporary Professor in Psychology and Ethology (Biology of Behavior) at the University of Paris (V, VIII & XIII). Since 1991, he is the Founder and Director of the Human Behavior Laboratory, University of Iceland, in formalized collaboration between 28 European and American universities based on “Magnusson’s analytical model” initiated at University René Descartes Paris V, Sorbonne, in 1995.
Abstract:
Striking similarity, but no self-similarity, exist between the mass-societies (i.e., of approximately >104 individuals) of such distantly related organisms as social insects and modern humans, while striking self-similarity exists between their structured mass-societies and those of their citizens that are mass-societies of cells that again are mass-societies of proteins (also called Cell Cities). Natural, as opposed to mathematical, self-similarity should probably be expected in a generally (statistical pseudo) fractal universe. This may be exemplified by the statistically self-similar repeated pattern type, called T-Pattern that is also characterized by significant translation symmetry, as it has been detected among other in the dynamic behavior and interactions of humans and in networks of neurons in living brains, while resembling (spatial) patterns on DNA. The brain-less mass-societies of proteins may thus provide useful and even essential ideas for the understanding of the biologically recent and first and only large-brain mass-societies, mostly evolved at the speed of cultural evolution among brains biologically evolved during a far longer (nomadic and illiterate) small-group “everybody-knows-everybody” past. Structural and functional (self-)similarities between protein and human mass societies are explored among others in terms of T-patterns appearing across time and space and from protein to human mass-societies, where extensive copying and spreading of standard T-patterned strings (texts) existing and evolving outside brains, seem reflections of the social ways of proteins, much as human schools, laws, religions and money.
